

Next: Dynamic Memory Allocation
Up: gendrift
Previous: Pointers and Arrays as
This type of simulation runs kind of slow. Let's start to use
optimization flags (-O) when you compile programs. When
compiler is translating the source code to object code, there are many
ways to translate the same source code. Compiler analyze the source
code, and it will try to make the fastest object code automatically
(optimization).
gcc -O3 source.c
There are several levels of optimization (-O1 -O2 -O3). Higher
number after -O means more optimization (potentially faster).
By default, gcc doesn't do any optimization (equivalent to -O0,
'Oh Zero').
You can see how the optimization influence the speed by
time ./a.out
- Make a function int CntMember(int arr[], int arrSize, int
member), which count the occurence of member in an array
arr of size arrSize and return the count.
- Use this CntMember() function to check the state of the
population every time after the next generation is created. You can
count the occurence of ``1'' and if the count becomes 0 or total
number of alleles in the population, the extinction or fixation of
the allele occured.
- Modify the code, so that it will run the simulations many times
(say 1000 replications), and confirm that the fixation probability
is equal to the initial frequencies.
- Now you want to collect data of how many generations the two
alleles will remain segregating (i.e. time to loss or fixation of 1
allele). Modify the source code to get this data.
Given the initial frequency of p, the average time to loss or
fixation can be approximated by
Kimura and Ohta (1969, Genetics 61:763) used diffusion
approximation. Compare your simulation results with this approximation
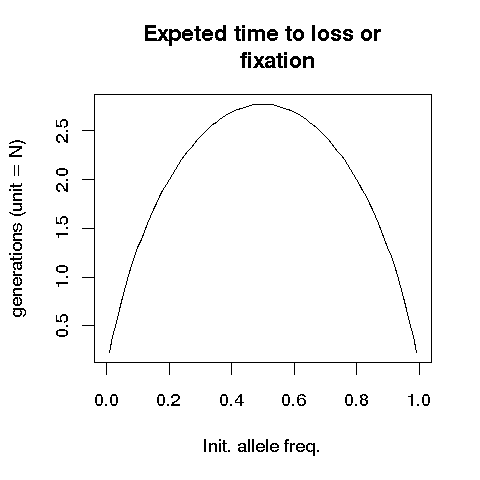
It remains segregating for the longest time when the initial
frequency of an allele is 1/2. This is approximately 2.77 N
generations.
- Consider that the population array represents the spatial
positions of a linear population (like plants growing along a road).
Previously, we assumed that the population is randomly mating, but
dispersion may be limited in real populations (``viscous''
populations). Modify the program to incorporate the limited
dispersal. Some parameter(s) should be controlling the
``viscosity'' (dispersal distance), and see how these parameters
influence the time to fixation or loss. You can make a function
CreateNextGenViscous().
- Selection vs drift
- We simulated a haploid population. Can you convert the simulation
to diploid case? You can cosider that 2 neighboring alleles in the
population array is a genotype of one diploid individual. In other
words, popB[0] and popB[1] are the two alleles of the
individual 1, and popB[2] and popB[3] are the two
alleles of the individual 2.
- Let's make that the two alleles are under selection. Fitness of
genotypes are:
| genotype |
0 0 |
0 1 |
1 1 |
| fitness |
1 |
1 + h s |
1 + s |
s is selection coefficient, h is dominance coefficient.
For example, you can consider that one beneficial mutant (allele
1) arose in the population (so the rest of the alelles are 0).
Even if the advantage of mutation is big, it will get lost most of
the time. You can check how the probability of ultimate fixation
of allele 1 is influenced by dominance.
- Other modification exercises:
- You can create two (or more) populations: each population
can be represented by an array. You can easily simulate gene
flow between the populations. Example, when you are creating
the next generation, you can draw a uniform random number [0,1)
and if it is less than probability of migration m, you
can draw the gamete from the other population(s).
- Instead of 2 alleles, you can have many alleles like
microsatelites. You can assume some mutational models (e.g,
allele 6 can only mutate to allele 5 or allele 7 etc).


Next: Dynamic Memory Allocation
Up: gendrift
Previous: Pointers and Arrays as
Naoki Takebayashi
2008-04-15
