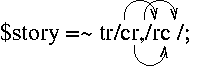- Matching operator
my $s = "scatter";
if ($s =~ /cat/) { ... } # true if $s contains "cat"
if (/dog/) { ... } # true if $_ contains "dog"
- Example of matching
#!/usr/bin/perl -w
$_ = "I Wish I Was a Mole In the Ground"; # using the special variable $_
# to simplify the comparison
print "Matched 1\n" if(/mole/); # no match
print "Matched 2\n" if(/e I/); # matches
print "Matched 3\n" if(/eI/); # no match
print "Matched 4\n" if(/Ground /); # no match
- Substitutions
$s =~ s/cat/wea/; # Replace cat with wea
Now $s contains "sweater".
As usual, the substitution operator s/ / / operates on default
scalar variable $_ if $varName =~ part is omitted:
s/cat/wea/;
- Simple example: DNA to RNA
#!/usr/bin/perl -w
my $seq = "ATTATGCGGCG";
print "DNA: $seq\n";
$seq =~ s/T/U/;
print "RNA?: $seq\n";
$seq =~ s/T/U/g;
print "RNA: $seq\n";
exit;
Global substitution. By default, s/ / / substitute only
the first match. s/ / /g will change the behavior, and ALL
instances which matches the pattern get substituted (g for global match).
- Transliteration
Try the following:
my $story = "My,cat,ate,a,rat!";
print "Before: $story\n";
$story =~ tr/cr,/rc /;
print "After: $story\n";
$story =~ tr/a-z/A-Z/;
print "Capitalized: $story\n";
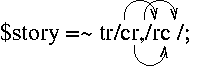
Useful when you want to take a string and replace every instance of
some character with some new character.
Does the following s/ / /g do the same thing as the tr/cr,/rc / above?
my $story = "A,cat,ate,a,rat!";
print "$story\n";
$story =~ s/c/r/g;
$story =~ s/r/c/g;
$story =~ s/,/ /g;
print "$story\n";
- We are reading in a file, and trying to find a line which is
exactly ``bert''. Does the following code do the job?
while(<IN>) {
chomp;
print "Found bert in line $.\n" if (/bert/);
}
It will find ``dogbert'', too.
- We can use anchoring metacharacter.
\A (^) means match at the beginning of the string
\z ($) means match at the end of the string
/\Abert/ # does not match "dogbert", but matches "bertram"
/^bert/
/bert\z/ # does not match "bertram", but matches "dilbert"
/bert$/
/\Abert\z/ # ONLY matches with exactly "bert"
/^bert$/
Note that the caret (^) has a different meaning now, since the caret is
not immediately after left square bracket.
- Improved version
while(<IN>) {
chomp;
s/\A\s+//; # remove leading spaces
s/\s+\z//; # remove trailing spaces
print "Found bert in line $.\n" if (/\Abert\z/);
}
If a line contains leading spaces (" bert") or trailing spaces
("bert "), the pattern (/^bert$/) won't match the
line. This kind of problem happens frequently, so it is a good idea
to clean up these leading/trailing spaces with the substitutions.
- Exercise:
Create a program, which reads in a file given in the command line
argument and removes empty lines (i.e. print out the
non-empty lines to the screen).
- The grouping character ( ) have an additional feature. You can
use it to extract a part of matching expressions.
- For each grouping, the part that matched inside get assigned to
special variables $1, $2, $3 etc.
# extract hours, minutes, seconds
if ($time =~ /(\d{2}):(\d{2}):(\d{2})/) { # match hh:mm:ss format
$hours = $1;
$minutes = $2;
$seconds = $3;
}
# Extract the sequence name from the FASTA sequence name line
if (/^\s*>\s*(.+)\s*$/) {
$seqName = $1;
}
- How do we know which parentheses corresponds to the 1st,
2nd,etc.
# extract hours, minutes, seconds
if ($time =~ /(((\d{2}):(\d{2})):(\d{2}))/) { # match hh:mm:ss format
$hms = $1;
$hm = $2
$hours = $3;
$minutes = $4;
$seconds = $5;
}
Just count the order of the opening parenthesis.
- Reusing the match within a regular expression
Instead of $1, $2, etc., you can use \1, 2, etc.
$_ = "abba";
if (/(.)\1/) {
print "Matched char: $1\n";
}
$_ = "ding dong";
if (/(.).(..) \1.\2/) {
print "$_ matched:$1:$2\n";
}
- Exercises:
- Create a program which checks if there are any micro-satelites
in a sequence. Let's say you are looking for repeat of 3 bases, and
there should be at least 5 repeats (e.g. AATAATAATAATAAT or longer).
- Modify the previous program to exclude the repeats are not
monomers (e.g. AAAAAAAA...).
- Create a program which will check if a protein sequence contain a
palindrome, which is at least 7 letter long. e.g., "RACECAR"
- When you download sequences from GenBank
in FASTA, they have
long names.
An example of sequence name:
>gi|49188826|gb|CO267808.1|CO267808 151H8 Opium poppy leaf cDNA library Papaver somniferum cDNA clone 151H8 5' similar to Unknown function, mRNA sequence
The accession number is red and italicized.
- You can use the following script to shorten the name lines
to accession numbers.
- Code
#!/usr/bin/perl -w
# shortenName.pl
# shorten the sequence names to accession numbers from fasta file.
while (<>){
chomp;
unless (/^>/) { # Not the name line
print "$_\n";
next;
}
# We are dealing with the name when the process reaches here.
s/^>\s*//; # get rid of '>'
my @line = split /\s+/;
my $first = shift (@line); # gi|49188826|gb|CO267808.1|CO267808
my @numbers = split /\|/, $first;
$accNum = $numbers[3];
$accNum =~ s/\.\d+$//; # remove version numbers
print ">$accNum\n";
}
exit;
- Wow, isn't it pretty cool that you can achieve the goal with
this short code? Let's take a look at a couple points in detail
- Diamond operator (<>).
#!/usr/bin/perl
while(<>) {
...
}
This is a short hand way of doing something similar to:
#!/usr/bin/perl
while (my $file = shift @ARGV) {
open (IN, "$file") || die "can't open $file\n";
while ($_ = <IN>) {
...
}
close (IN);
}
This kind of operations happens frequently (Parsing many files, line
by line). Instead of writing out all of these, you can use while(<>){ }. This operator is as valuable as diamonds. Perl
programmers love to be lazy!
- Simpler example:
#!/usr/bin/perl
# kitty.pl
while(<>) {
print $_;
}
If you do,
bash$ kitty.pl file1.txt file2.txt file3.txt > output.txt
the three files will be concatenated into one output.txt file. This
kitty.pl works like the unix command cat.