my @a = (1,2,3,4,5); my $first = shift (@a); # $first = 1, @a = (2,3,4,5) unshift (@a, $first); # $first = 1, @a = (1,2,3,4,5) my $last = pop (@a); # $last = 5, @a = (1,2,3,4) push (@a, $last); # $last = 5, @a = (1,2,3,4,5)
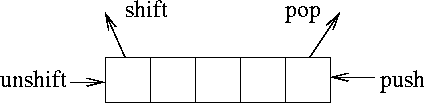
In addition to removing an element from the list, shift() and pop() returns the removed element. So you can assign the removed element to another variable (e.g. $first or $last above) if needed. These two function returns a special value undef if an empty array is given.
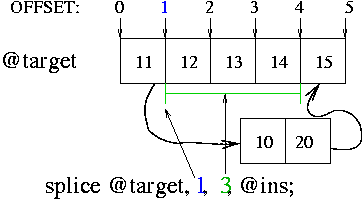
# splice @targetArray,OFFSET,LENGTH,@insertArray my @target = (11,12,13,14,15); my @ins = (10, 20); splice @target, 1, 3, @ins; # @target becomes (11, 10, 20, 15), @ins unchanged

@target = (11,12,13,14,15); splice @target, 3, 1; # @target = (11, 12, 13, 15) splice @target, 0, 1; # @target = (12, 13, 15), same as shift(@target) splice @target, -1, 1; # @target = (12, 13), same as pop(@target)
splice @target, 0, 0, (10, 11) # @target = (10,11,12,13)
# same as unshift(@target, (10, 11));
splice @target, @target, 0, 15 # @target = (10,11,12,13,15)
# same as push(@target, 15);
splice @target, 4, 0, 14 # @target = (11,12,13,14,15)
@removed = splice @target, 0, 4; # @removed contains the first 4 elements.
my @a = (1,2,3); my @b = reverse(@a); # @a = (1,2,3), @b = (3,2,1)
It returns an array whose order is reversed. Note that the argument array (@a) is unaltered. If you want to reverse an array ``in place'', you can assign it back into the same variable:
@a = reverse(@a); # @a = (3,2,1) now.
You can also use reverse to character strings (scalar variable).
my $s = "hot dog"; my $rev = reverse($s); print $rev, "\n"; # becomes "god toh";
@fileContent = <INFILE>; # ("line1\n", "line2\n","line3\n", ...)
chomp(@fileContent); # ("line1", "line2","line3", ...)
Then it removes newline from each element.
my @x = sort("small", "medium", "large"); # @x is ( "large", "medium","small")
my @y = (1,2,4,8,16,32,64);
@y = sort(@y); # @y = (1,16,2,32,4,64,8)
@y = sort by_numerically (@y); # now @y = (1,2,4,8,16,32,64);
sub by_numerically {
if ($a < $b) {
return -1;
} elsif ($a == $b) {
return 0;
} elsif ($a > $b) {
return 1;
}
}
| -1 | If value of $a is considered to come eariler than the value of $b in your custom ordering scheme |
| 0 | If values of $a and $b is equivalent |
| +1 | If value of $a is considered to come later than the value of $b |
@y = sort {$a <=> $b} (@y); # now @y = (1,2,4,8,16,32,64)
<=> operator does the exactly same thing as sort_numerically().
my @x = sort by_rev ("small", "medium", "large");
sub by_rev {
my $rA = reverse $a;
my $rB = reverse $b;
if ($rA gt $rB) {
return -1;
} elsif ($rA lt $rB) {
return 1;
} else {
return 0;
}
}
@x = qw(small medium large); # equivalent to @x = ('small', 'medium', 'large');
If you are initializing a large array or hash with character strings,
it become tedious to type in quotes for each element. You can use
qw to split it by spaces, and put quotes around each element.
An example of initializing a large hash (codon -> amino acid):
# '*' indicates the termination codon
%aminoAcid = qw (TTT F TTC F TTA L TTG L TCT S TCC S TCA S TCG S TAT Y TAC Y TAA * TAG * TGT C TGC C TGA * TGG W CTT L CTC L CTA L CTG L CCT P CCC P CCA P CCG P CAT H CAC H CAA Q CAG Q CGT R CGC R CGA R CGG R ATT I ATC I ATA I ATG M ACT T ACC T ACA T ACG T AAT N AAC N AAA K AAG K AGT S AGC S AGA R AGG R GTT V GTC V GTA V GTG V GCT A GCC A GCA A GCG A GAT D GAC D GAA E GAG E GGT G GGC G GGA G GGG G);
my @a = ("See", "you");
my @b = ("later", "Aligator");
# method 1 (better)
push (@a, @b);
print "@a\n";
# method 2 (slower)
@a = (@a, @b);
Perl doesn't have nested arrays (an array as an element of another array). In other words, (("a1", "a2"), ("b1", "b2")) automatically becomes ("a1", "a2", "b1", "b2"). So the 2nd method will produce a one dimensional array (automatic flattening).
But push is a more efficient way to achieve the same goal.
my @a = (1,3,5,2,5,4,3,2,1,5);
my @uniqArr = Unique(@a); # you get (1,2,3,4,5)
sub Unique {
my %seen =();
foreach my $element (@_) {
$seen{$element}++;
}
return (sort(keys(%seen)));
}
@matched = ();
foreach my $i (@list) {
push (@matched, $i) if ($i =~ /^\d+$/ && $i < 20);
}
This will find all integers which are less than 20.
An easier method:
@matched = grep {$_ =~ /^\d+$/ && $_ < 20} @list;
Each element gets assigned to $_ and if it satisfy the test (inside of { }),
the value gets inserted to @matched.
Let's say we have a hash table %age, which contains the age of each person (the keys are names of people). Can you write a grep statement to get an array of names (keys in this hash), whose age is younger than 21?
my $song = "Old Joe Clark"; $song = lc($song); # become "old joe clark" $song = uc($song); # become "OLD JOE CLARK" $song = lcfirst($song); # become "oLD JOE CLARK" $song = ucfirst($song); # become "OLD JOE CLARK"Note that lcfirst and ucfirst only change the first character of string, and the case of the other characters are not changed.
Can you make a function which take a string as the argument, and
return a string with the first character of each word is upper case,
but the rests are lower case? So Capitalize($song) will always
return "Old Joe Clark".
my $string = "kermit the frog"; my $len = length($string); # 15 characters including spaces in $string
@line = split /\s+/, $_; @line = split /\s+/; @line = split; @csvdata = split /\s*,\s*/, $csvString;The function split the string (2nd argument) by the pattern inside of / /, and return an array. Note that we are using regular expressions now.
If you omit the 2nd argument (string scalar), it operates on $_. If you omit the pattern, it splits on whitespace (after skipping any leading whitespace). So the first 3 statements are equivalent.
@a = (1, 20, 36)
print join("\t", @a), "\n" # print out a tab delimited line "1\t20\t36".
my $concatenated = join("", @a) # $concatenated is 12036
my ($hour, $min, $sec) = (20, 31, 16);
my $timeString = join (':', $hour, $min, $sec); # become 20:31:16
my $sep = "\n";
$threeLine = join($sep, @a);
Note that split() uses a pattern / /, but join takes a character string.
If you do:
uniqAcc.pl fileA fileB > outFile
the result should look like the following outFile
fileA: AY167979 Bras.juncea # Brassica juncea rbcL gene AY167976 Bras.rapa # Brassica rapa rbcL AF267640 Bras.napus # Brassica napus rbcL fileB: AF267640 B.napus # Brassica napus rbcL AY167979 B.juncea # Brassica juncea rbcL gene U91966 A.thal # Arabidopsis thaliana rbcL outFile: AY167979 Bras.juncea # Brassica juncea rbcL gene AY167976 Bras.rapa # Brassica rapa rbcL AF267640 Bras.napus # Brassica napus rbcL U91966 A.thal # Arabidopsis thaliana rbcL
Reverse complement of 'AAGCTTGC' is 'GCAAGCTT'.
selectSites.pl in.fasta 6 200
It will read in the fasta file, and select sequences between site 6 and site 200 from each sample, and print out the FASTA file.