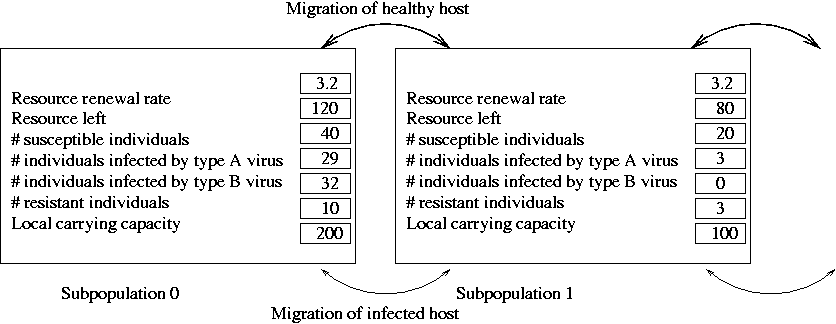
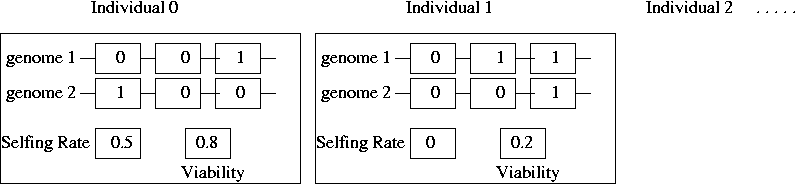
- Structure can contain other structures as the members.
struct line {
struct coord begin;
struct coord end;
};
struct line l1;
l1.begin.x = 0;
l1.begin.y = 0;
l1.end.x = 10;
l1.end.y = 10;
- Arrays and pointers can be the members of structures.
struct genotype {
int locus1[2];
int locus2[2];
} g1;
g1.locus1[0] = g1.locus1[1] = 1;
struct myArray {
double *data;
int size;
} arr;
arr.data = (double *) calloc(20, sizeof(double));
arr.size = 20;
- Structure can be passed to a function
CntLessThan(arr, 10.0);
int CntLessThan(struct myArray thisArr, double x) {
int i, cnt = 0;
for (i = 0; i < thisArr.size; i++) {
cnt += (thisArr.data[i] < x);
}
}
- pointers to structures.
struct myArray *aPtr;
aPtr = & arr;
(*aPtr).data[0] = (*aPtr).data[1] + (*aPtr).data[2];
Don't forget the parenthese here.
- Using a pointer to the structure with the indirect membership operator (->)
The member of a structure pointed by a pointer can be also accessed with ``->''
tmp = aPtr->size - 1;
tmp = (*aPtr).size - 1;
These two statements are exactly same.
- You can create an array of structures
struct genotypes pop[100];