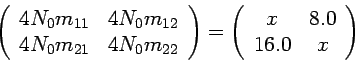

Next: Approximate Bayesian Computation
Up: coalsim
Previous: Coalescent
Subsections
Perl is frequently used to write custom analysis pipelines, which
drives other programs and makes automated analyses.
Here we will look at how to write perl scripts which interact with
other programs.
- You can give any unix commands you can execute in the command
line (shell) with system().
#!/usr/bin/perl -w
system("date");
my $cmd = "ls";
my $opt = "-l";
my $arg = "/";
system("$cmd $opt $arg");
exit;
The output of the shell commands will go to the output of the perl
script. By default, it is the screen, i.e. STDOUT, but you can
redirect it with > (e.g. shellCmds.pl > output).
- Be careful when your command contains shell environmental
variables (e.g.
$PATH).
system('echo $PATH'); # Works with single quotes
system("echo $PATH"); # Perl thinks that $PATH is a perl var
# because " " causes interpolation.
system("echo \$PATH"); # Escaping $ works.
- Exit status of shell commands:
- Several summary statistics (we talked about
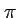 , S, Tajima's
D) can be calculated by another program sample_stats,
which is distributed with ms.
, S, Tajima's
D) can be calculated by another program sample_stats,
which is distributed with ms.
ms 30 4 -t 3.0 | sample_stats
prints out:
pi: 4.232 ss: 13 D: 0.956 thetaH: 3.285 H: 0.947
pi: 1.314 ss: 5 D: 0.114 thetaH: 0.478 H: 0.836
pi: 2.540 ss: 8 D: 0.783 thetaH: 1.942 H: 0.597
pi: 2.726 ss: 14 D: -0.762 thetaH: 1.549 H: 1.177
These different summary statistics represents different aspects of
genealogy which created the sequence data.
- Exercises:
Create a perl script (cleanSampleStats.pl), which will read in the output of sample_stats,
and convert the format to a nice tab-delimited numbers. i.e., the
output should look like:
pi: ss: D: thetaH: H:
4.232 13 0.956 3.285 0.947
1.314 5 0.114 0.478 0.836
2.540 8 0.783 1.942 0.597
2.726 14 -0.762 1.549 1.177
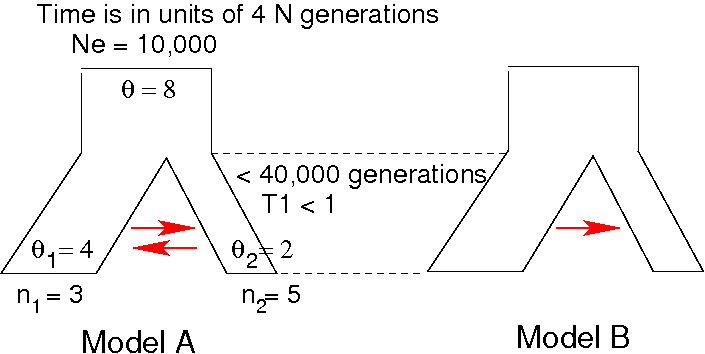
Let's say we have sequence data from two populations (e.g. Alaska
mainland and Kodiak Island).
We are wondering whether the migration is one directional.
We want to run coalescent simulations under two different models, and
look at the results of simulation to see whether the observed sequence
data fits one of the model better.
Instead of setting the migration rate to a pre-fixed value, modify the
program to estimate the migration rate (in addition to
divergence time). In other words, you need to draw the migration from
a random distribution.


Next: Approximate Bayesian Computation
Up: coalsim
Previous: Coalescent
Naoki Takebayashi
2011-11-09